|
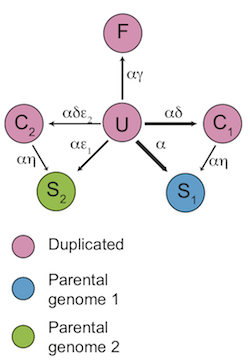
I have also studied why bakers' yeast is able to turn sugar into alcohol with such efficiency. Although we still do not understand all the details of this story, genes that produce ethanol have been duplicated in the yeast genome through a whole-genome duplication (WGD). We think these duplications contribute to the process of evolving new functions in two important ways. Firstly, it is sometimes enough to have a second copy of a gene to obtain a new function. The reason is that genes are the templates from which the machinery of the cell is made. Having an extra copy of a gene can therefore allow more copies of a given protein to be made, potentially allowing the cell to grow faster. Perhaps more importantly, these extra copies of genes can be altered such that they perform a new function that is related to their original function, much as computer users often open an old letter and then modify it to make a new one. In addition to gene duplication, bakers' yeast has also changed in other ways: the timing with which certain genes are turned on and off has also been altered, allowing yeast to more closely control how it uses the sugar it finds in its environment. The result is an evolutionary advantage based on the fast, inefficent use of glucose.
|
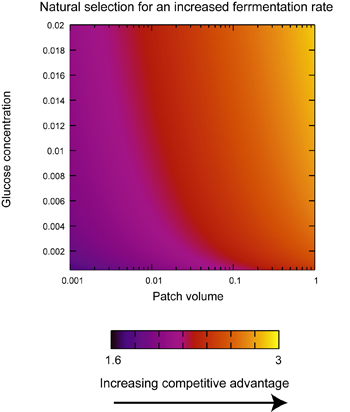
Simulations of competition between slow and fast fermenters |

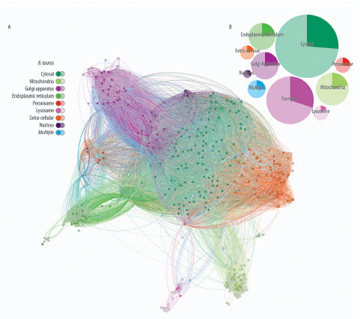

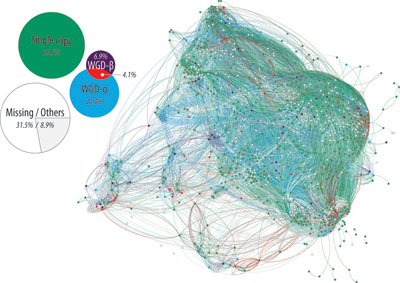
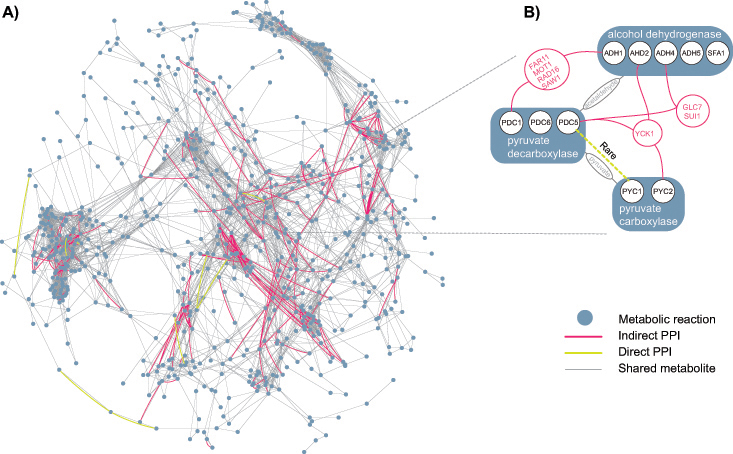 In this picture, the grey lines join enzymes (blue dots) that use the same metabolites. The superimposed red lines are cases where those two proteins are also connected by a third protein, that is not an enzyme (see right).
In this picture, the grey lines join enzymes (blue dots) that use the same metabolites. The superimposed red lines are cases where those two proteins are also connected by a third protein, that is not an enzyme (see right).